By Mathieu Boudreau
The April 2020 MRM Highlights Reproducible Research Insights interview is with Jieun Lee and Jongho Lee, researchers at Seoul National University in South Korea, and authors of a paper entitled “Artificial neural network for myelin water imaging”. This paper was chosen as the MRM Highlights pick of the month because it reports good reproducible research practices. In particular, in addition to sharing their code, the authors also shared their trained deep learning networks. For more information about Jieun and Jongho and their research, check out our recent interview with them.
General questions
1. Why did you choose to share your code/data?
Our main reason for sharing our code is to allow readers to directly access the materials used in the paper so that they can apply it to their research. As regards deep learning research, we know from experience that describing the neural network architecture is not sufficient if the aim is to produce reproducible research, because the training data and network configuration used to train the deep learning model can substantially change the performance of the network. Therefore, we decided to share not only the code, but also the pre-trained network. This means that future studies can compare their network performance with that shown in our study.
2. Is this a regular habit of your lab? Or, if this is the first time you have done it, do you think you’ll share code/data again in future publications?
Yes, sharing code and software tools is common practice in our lab. We have already shared lab-developed softwares (pulse sequences and reconstruction algorithms) on our webpage. For our deep learning projects, we created a GitHub lab organization dedicated to host them. So far, we have our QSM (QSMnet), myelin water imaging (ANN-MWI), and RF design (DeepRF_SLR) deep learning projects on GitHub. We intend to continue sharing our code in future publications.

3. At what stage did you decide to share your code/data? Is there anything you wish you had known or done sooner?
Since sharing code has become a default practice in our lab, we decided to share the code for this project in the early stages of writing the paper. We uploaded the code immediately before submitting the paper to Magnetic Resonance in Medicine.
4. Are there any other reproducible research habits that you didn’t use for this paper but might be interested in trying in the future?
In addition to the code, we also wanted to share the data used to train and test the models. However, our institutional review board (IRB) policies on privacy made it difficult to share the data. Basically, we have to obtain consent signatures from subjects prior to sharing data. Unfortunately, our data were drawn from previous studies and were a few years old, therefore we were unable to get the required signatures. Now, we have a revised IRB policy on data sharing that allows us to share datasets from our current projects in future studies.
Questions about the specific reproducible research habit
1. What advice do you have for people who would like to share their deep learning code/data along with their paper?
For deep learning, we believe that it is important to share not only the code, but also the pre-trained network and data, because the network performance depends on the network structure, its initialization, and the training data set itself. If these are not available, it is difficult to test the reproducibility of the method and to perform a comparison between methods. One option for data sharing is to develop a public dataset, such as the one for parallel imaging done very nicely by the NYU group.
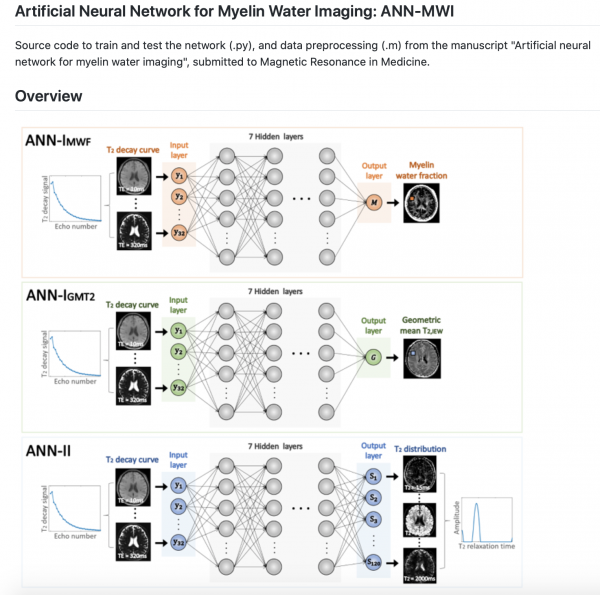
2. You shared some trained models along with your deep learning training code. What are the benefits (and limitations) of sharing the trained models instead of the raw data?
As mentioned before, we hoped to share the data but were unable to do so due to the restrictions imposed by the IRB. Sharing the code and data is not enough to guarantee reproducibility because the initialization of the network affects its performance too. Hence, you need to share a complete set: code, data and pre-trained network (or initialization).
3. How do you recommend that people use the project repository you shared? Can they use the trained model you shared as-is, or should they generate their own training datasets?
You can use our trained model as-is, or use it as a seed model to do transfer learning with your own network. The latter will be helpful in accelerating the training speed of a network and could help you use a smaller data size for training.
4. Could your trained model be adapted to another experiment?
It depends on the experimental setup (sequence, scan parameters, patient type, etc.). For our network, we used the data from a popular sequence and commonly used scan parameters for T2-based myelin water imaging, precisely so that the network might be used for other studies. However, with a different setup it may or may not be possible to use our network. For example, if your data were acquired using different echo spacings from ours, the performance will degrade (as we demonstrated in our paper). This issue of network generalization is an important topic in deep learning, and it is one that we are currently exploring.




