By Mathieu Boudreau
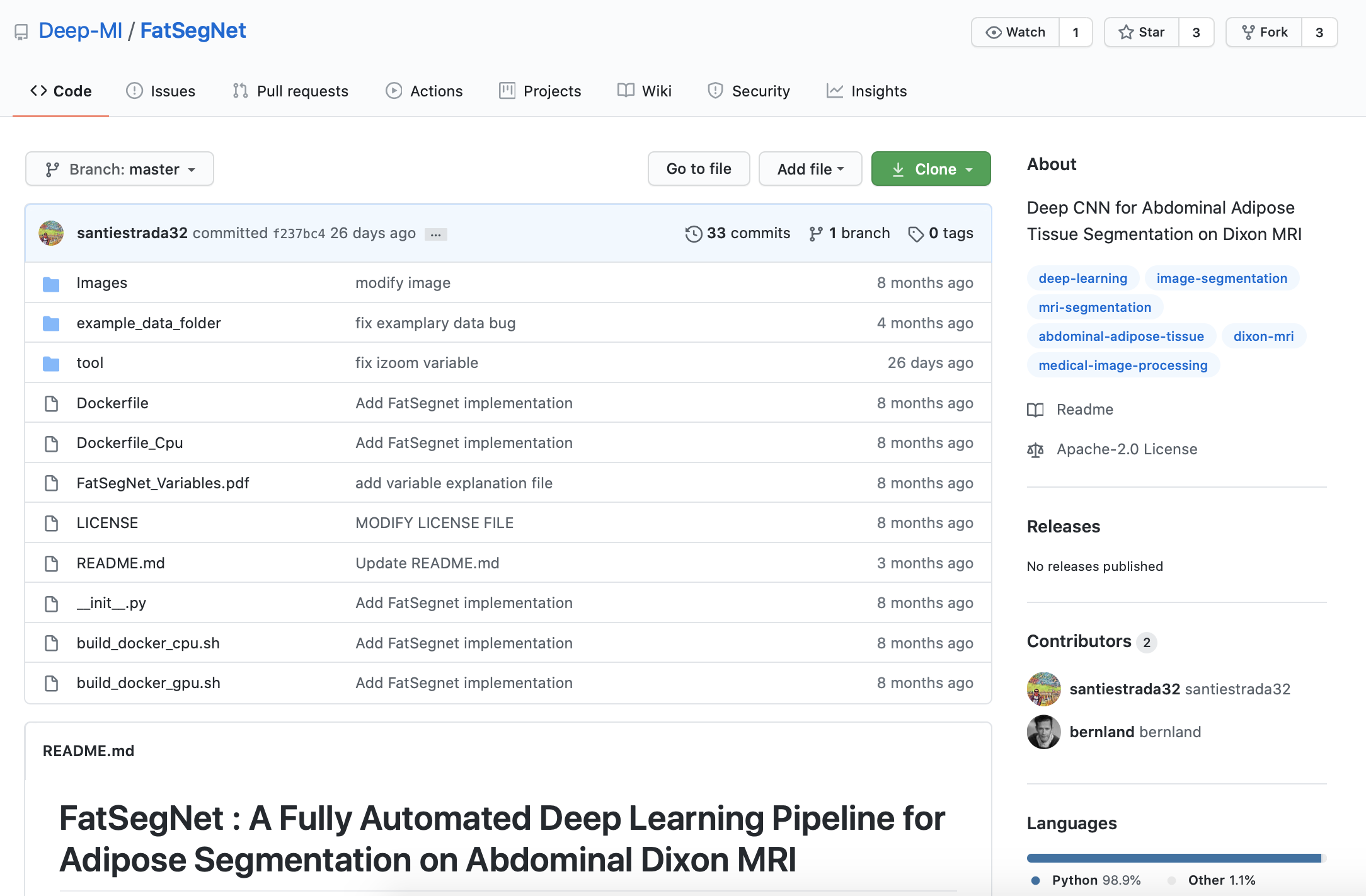
The June 2020 MRM Highlights Reproducible Research Insights interview is with Santiago Estrada and Martin Reuter, researchers at the German Center for Neurodegenerative Diseases in Bonn, Germany, and the A.A. Martinos Center for Biomedical Imaging in Boston, and authors of a paper entitled “FatSegNet: A fully automated deep learning pipeline for adipose tissue segmentation on abdominal Dixon MRI”. This paper was chosen as the MRM Highlights pick of the month because it reports good reproducible research practices. In particular, in addition to sharing the code related to FatSegNet, the authors also shared Dockerfiles enabling others to reproduce the coding environment needed to run their software. For more information about Santiago and Martin and their research, check out our recent interview with them.
Crash Course: Dockerfiles
 Dockerfiles are scripts that provide the necessary instructions to create Docker images, which are reproducible and shareable computing environments. Unlike Docker images, which can be up to several gigabytes in size and are compiled, Dockerfiles are lightweight and can be version controlled with softwares like git. Creating a Dockerfile can be fairly easy if you inherit a base Docker image that has already been generated with most of the softwares and setup you need (i.e. Python, Jupyter, etc); these are hosted on Dockerhub and are called within Dockerfiles with the “FROM” command. For example, FatSegNet users inherit a TensorFlow Docker image in their Dockerfile, so they don’t need instructions on installing TensorFlow – it’s already done for them. You can add additional instructions using the “RUN” command (i.e. to install additional packages and softwares into your environment, as is done here for FatSegNet). There are several other commands that can help you in setting up the environment the way you want, like sharing a folder between your computer and the Docker container. Loads of resources are available online to help you with your journey into the realm of Dockerfiles – here are 10 tips for some best practices.
Dockerfiles are scripts that provide the necessary instructions to create Docker images, which are reproducible and shareable computing environments. Unlike Docker images, which can be up to several gigabytes in size and are compiled, Dockerfiles are lightweight and can be version controlled with softwares like git. Creating a Dockerfile can be fairly easy if you inherit a base Docker image that has already been generated with most of the softwares and setup you need (i.e. Python, Jupyter, etc); these are hosted on Dockerhub and are called within Dockerfiles with the “FROM” command. For example, FatSegNet users inherit a TensorFlow Docker image in their Dockerfile, so they don’t need instructions on installing TensorFlow – it’s already done for them. You can add additional instructions using the “RUN” command (i.e. to install additional packages and softwares into your environment, as is done here for FatSegNet). There are several other commands that can help you in setting up the environment the way you want, like sharing a folder between your computer and the Docker container. Loads of resources are available online to help you with your journey into the realm of Dockerfiles – here are 10 tips for some best practices.
General questions
1. Why did you choose to share your code/data?
We believe in reproducible research and we think more open-source tools should be available to the scientific community for validation, replication and application. We hope that FatSegNet will allow researchers to segment adipose tissue in abdominal Dixon MR scans in a fast and reliable way.
2. Do you plan to share code/data regularly in future publications?
Yes, we plan to continue making our tools open-source. We will announce any new tools on our Twitter account @deepmilab and webpage https://deep-mi.org/, so simply follow us to stay up to date.
3. At what stage did you decide to share your code/data? Is there anything you wish you had known or done sooner?
From the beginning our plan was to make the tool open-source. We recommend considering this step early and thinking about the software license, as there are very different types. We recommend permissive licenses (e.g. MIT, Apache, BSD) as they allow wide use and also combination of methods into other large open-source software projects (best compatibility).
4. Are there any other reproducible research habits that you didn’t use for this paper but might be interested in trying in the future?
In the future, we would like to release some example cases for our tools so that people can test the code after installation. Currently, with FatSegNet, we cannot provide examples due to data protection restrictions. We are working towards increasing flexibility of data sharing as part of the informed consent, so that some cases can be released on open-source repositories.
Questions about the specific reproducible research habit
1. What advice do you have for people who would like to use Docker containers in their research ?
Try them. Docker images are really easy to run and to make. They eliminate the challenges of setting up your code to work in different environments. Your Docker image should run the same no matter where it is running.
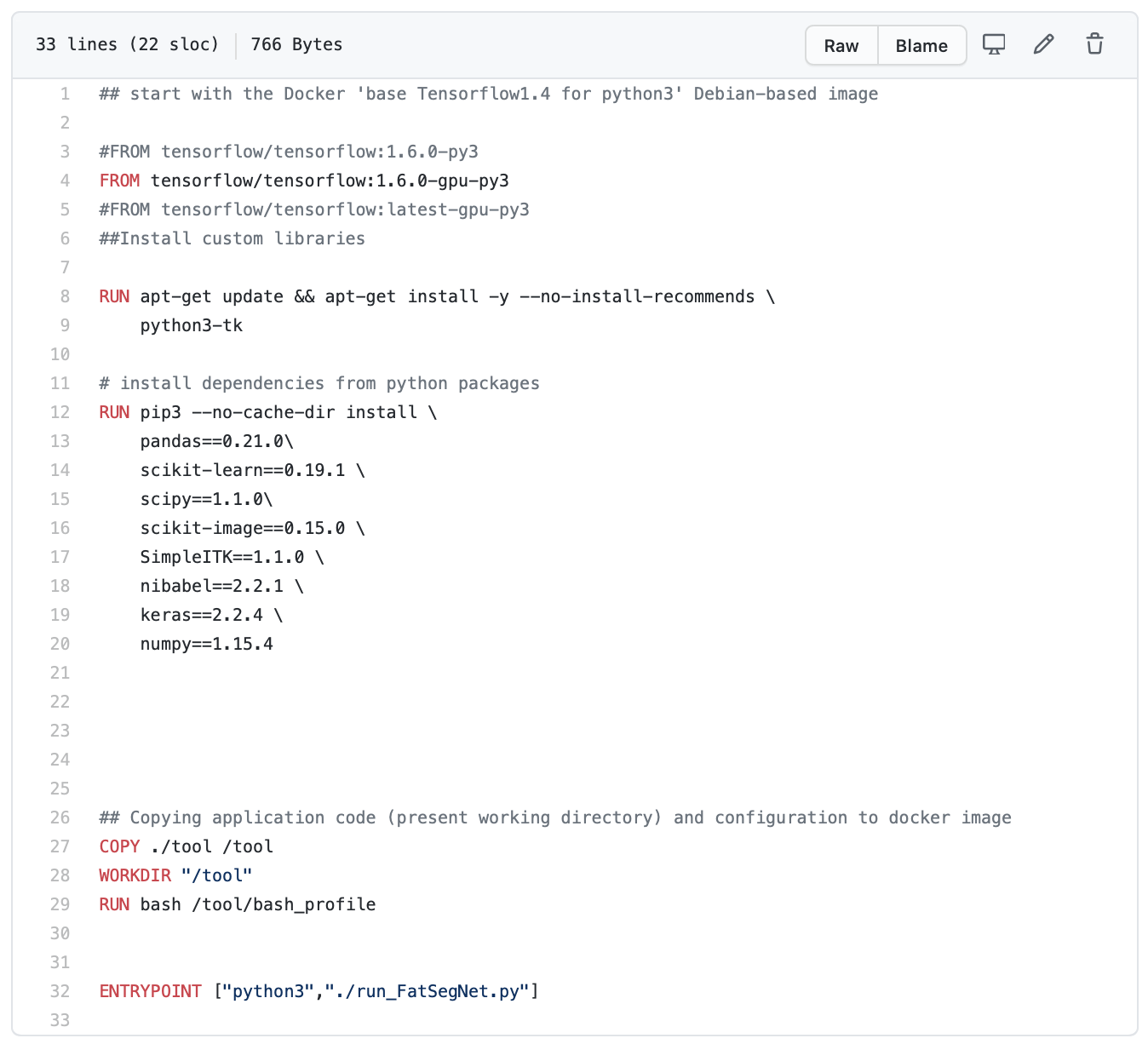
2. Can you share some resources to help our readers get started with using Docker?
Docker has a really easy online tutorial for those who want to learn the basics and Stack Overflow is a very good hub for asking questions and getting solutions to most problems you’ll encounter.
3. Did you come up against any challenges or hurdles working with Docker containers?
The biggest challenge was working with GPUs, however, the newer releases of Docker are more intuitive for using these types of resource. If you plan on using GPUs from within Docker, we recommend that you first check the following site: https://github.com/NVIDIA/nvidia-docker.
4. Do you get any other benefits from using Docker containers?
We like Docker because it allows the code to be portable and reduces installation incompatibilities. Frequently, open-source implementations cannot be used because they require a different library version or don’t compile with any modern compilers. With Docker you can create a lightweight image of your executable tool bundled with all necessary requirements and eliminate the “it works (only) on my machine” problem.




