By Mathieu Boudreau
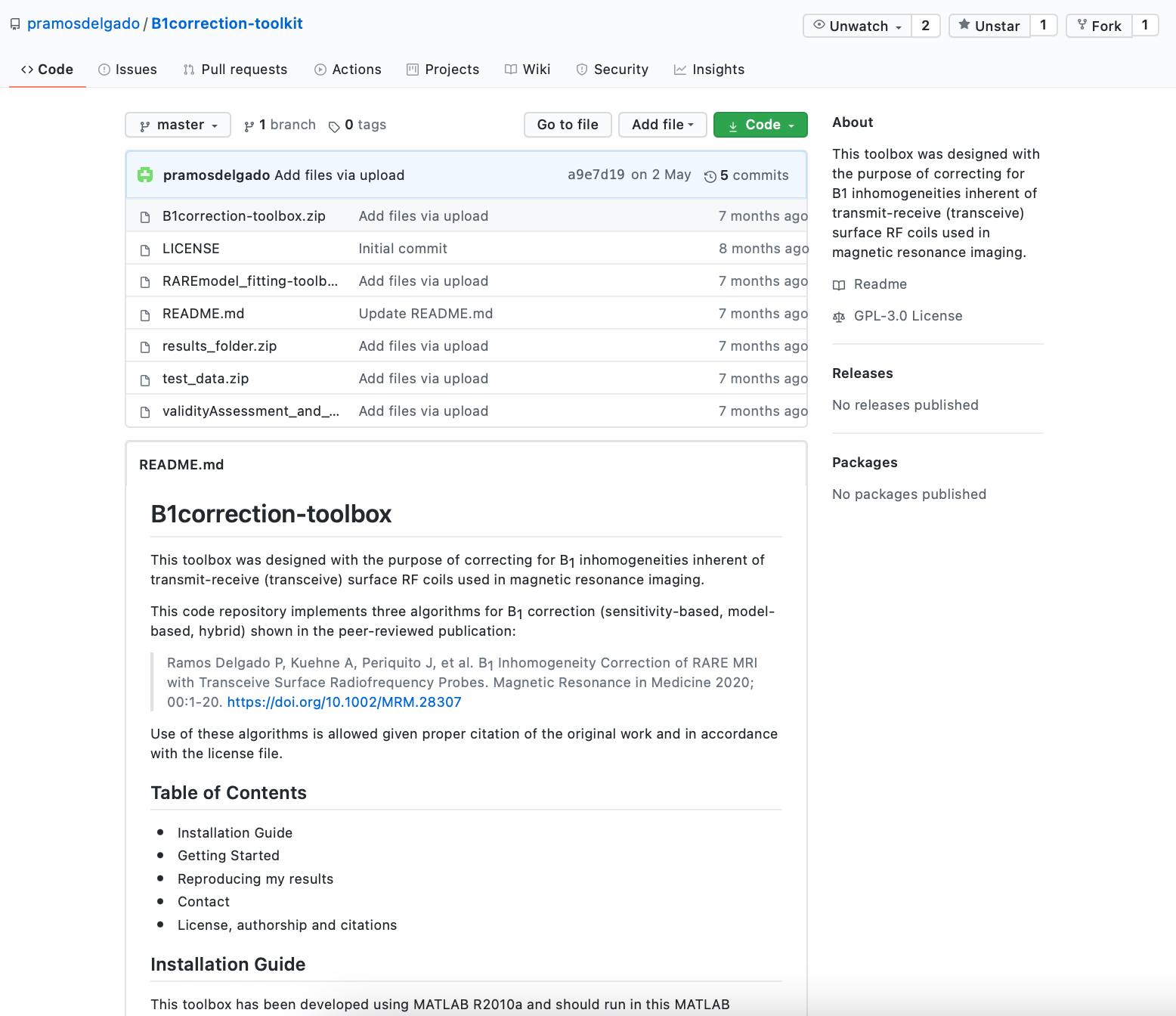
The November 2020 MRM Highlights Reproducible Research Insights interview is with Paula Ramos Delgado and Thoralf Niendorf, researchers at the Berlin Ultrahigh Field Facility (B.U.F.F) in Berlin, Germany, and authors of a paper entitled “B1 inhomogeneity correction of RARE MRI with transceive surface radiofrequency probes”. This paper was chosen as the MRM Highlights pick of the month because it reports good reproducible research practices. In particular, in addition to sharing their code and data, the authors also provided exemplary documentation in their GitHub software repository and inside their code. To learn more about Paula and Thoralf and their research, check out our recent interview with them.
General questions
1. Why did you choose to share your code/data?
Thoralf: For the purpose of advancing healthcare, we need to ensure reproducibility and broad dissemination of the outcome of our research. We therefore want to make sure that other people can easily access our work without coming up against any boundaries or paywalls.
Paula: I am a firm believer in sharing code and data to make the entire publishing process more transparent. I have lost count of the number of papers I have read that have left me thinking “Yes, you got beautiful results, but you are not really telling me how you actually did it, are you?” It used to be that scientists would tell you only as much as they needed to in order to get their papers published, and they would be quite vague in their descriptions, because they did not really want you to replicate their approach, or at least not without going through a lot of hassle first, as they wanted to feel they were keeping the know-how advantage. In the end, though, I think the whole “publish or perish” philosophy goes against sharing, which is a pity. But attitudes have been changing in the last few years. I saw a lot of special open-source sessions during the ISMRM meeting in Montréal. It was very exciting to see that the MR community is interested in sharing and making MRI data and code available openly. I think this is very important to advance the science and to help us realize that we need to stop wasting time by constantly re-inventing the wheel.
2. What is your lab or institutional policy on code/data sharing?
Thoralf: We encourage the sharing of code/data whenever we can. Yet, there is no institutional policy on this so far. Unfortunately, many journals do not yet offer a space for sharing data/code.
Paula: I’d say that we, in our group, are all pretty much in favor of sharing. Within the institute, I have become aware of more and more talk of reproducibility and open science lately. Also, here in Germany, we have a new agreement between Wiley and several German Institutions (Projekt DEAL) to publish Open Access for free, and we took advantage of this opportunity for our publication.
3. At what stage did you decide to share your code/data for this project? If it was late in the project, is there anything you wish you had known or done sooner? If it was early on, what did you do in preparation to do so?
Thoralf: We had already started the project when MRM entered the pilot phase for sharing code/data. As soon as we knew about the MRM initiative we decided to use this project as a role model for our future approach.
Paula: True. I had this idea of making the code open but imagined doing so after the paper was published. But when we submitted the manuscript, both the editor and the reviewers suggested that we provide the code and data with it. So, we thought it was perhaps valuable to do it during the review process (which fortunately was fairly painless!) and add a sentence within the paper giving the GitHub link, so that people would know where to find the repository.
4. How do you think we might encourage researchers in the MRI community to contribute more open-source code along with their research papers?
Thoralf: Encouragement is important, but I believe that journals should make data/code sharing mandatory.
Paula: I, too, think that sharing code and data (and even pulse sequences!) should be the norm and not the exception. It needs to be understood that when everyone contributes, everyone is a winner. That way, instead of being faced with black boxes, we could apply new techniques to our own data, and immediately focus on developing other ideas. Also, sharing code/data increases the credibility of a paper: it allows readers to see with their own eyes that the method is working just as you, the authors, claimed.
Questions about specific reproducible research habits
1. What advice do you have for people who would like to create well-documented code repositories?
Thoralf: We use GitHub, as it is convenient. Just sharing data/code is not enough, though. It should also be very well documented.
Paula: What I did was first make a list of all the scripts and data necessary to replicate the analysis and figures. I am a natural list-maker, and so this step came quite easily to me!
I also gave variables and datasets self-explanatory names so that they would be easy for others to grasp. I also extended the comments in the code so that it was easier for a reader to quickly understand what was going on.
And while I was doing this, I wrote, in parallel, a document in Word — this later became the README.md — that explained, step by step, how to run each of the scripts from the point of view of a total MATLAB newbie, and what you can expect to find in each dataset.

2. Can you share some resources to help the audience get started using GitHub and Markdown?
Paula: This was the first project I created in GitHub, and so I, myself, had to do a bit of reading first and create a new account. GitHub itself was easy to grasp, not much different from a cloud where you upload your files. Markdown was a bit more of a challenge because I had to learn how to design a proper and legible text format for the README from scratch. I basically googled “readme file GitHub” or something similar and found several interesting resources. It also helped give me more ideas on how to structure the README file itself.
3. What questions did you ask yourself while you were documenting your code?
Thoralf: Is it enough to share the code/data? Should there be a harmonized way of documenting the data/code for the sake of re-implementation and reproducibility by others?
Paula: See my answer to question 1) of this section.
4. Are there any other reproducible research habits that you haven’t used for this paper but might be interested in trying in the future?
Paula: One thing to note is that the code has recently been added to the OpenSourceImaging.org (OSI2) platform (see here), which is an initiative that we are co-founders of. We had some compatibility problems with some of the images, which is why there was a delay in uploading it to the platform. Also, I personally would like to start developing all my future code in Python. MATLAB is a proprietary software and I am aware that people need a license to be able to run the code we provided. Python and Jupyter Notebooks seem to be the way forward for documenting open-source code, in my opinion.





