By Mathieu Boudreau
This MRM Reproducible Research Insights interview is with Julien Songeon and Antoine Klauser from the University of Geneva in Switzerland, respectively first and last author of a paper entitled “In vivo magnetic resonance 31 P-Spectral Analysis With Neural Networks: 31P-SPAWNN”. This work was singled out because it demonstrated exemplary reproducible research practices; specifically, the authors shared the source code for their proposed method (simulations, their deep learning network, and sample datasets).
To learn more, check out our recent MRM Highlights Q&A interview with Julien Songeon and Antoine Klauser.
General questions
1. Why did you choose to share your code/data?
This approach naturally facilitates the demonstration presented in the manuscript, allowing interested readers to test the method themselves. Also, we enable others to validate and reproduce our results effectively.
Also, rather than adopting a competitive approach, we prefer, in this way, to promote collaboration in MR research, because a collaborative mindset can foster advancements and progress in the field. Ultimately, code sharing can assist other researchers who wish to pursue similar research directions. They can utilize our work as a foundation, potentially saving significant implementation and coding efforts. This accelerates the pace of research and encourages researchers to build upon existing knowledge rather than wasting resources on replication efforts.
2. What is your lab or institutional policy on sharing research code and data?
Our institution has a highly flexible policy regarding the sharing of code and data. While open science practices are strongly encouraged, there are no clear advantages for us nor mandatory constraints in place that require us to share .
3. How do you think we might encourage researchers in the MRI community to contribute more open-source content along with their research papers?
I believe that if code and data sharing were recognized to add clear value to a submitted manuscript, then authors would have a positive incentive to provide open-source content. In addition to evaluating factors such as innovation, soundness of methodology, quality of results, and clarity of demonstration, editors should acknowledge the inclusion of code and data as a valuable contribution to the scientific community.
Questions about the specific reproducible research habit
1. Your code repository is really well documented. What advice would you give to people preparing to share their first repository?
When preparing to share a code repository, you need to think about the future users of your code and what information they will need to understand and use it effectively. The README file should explain the purpose of the code, and provide installation instructions, dependencies, and examples. The files could have comments within the code to clarify specific sections or functions. To help users navigate the repository and locate relevant files easily, I suggest organizing your code and files in a logical manner, using explicit naming. Finally, I would recommend including examples and/or tutorials, providing sample input data and scripts that demonstrate how to use your code. This can really help users to understand the expected workflow and apply the code to their own research.
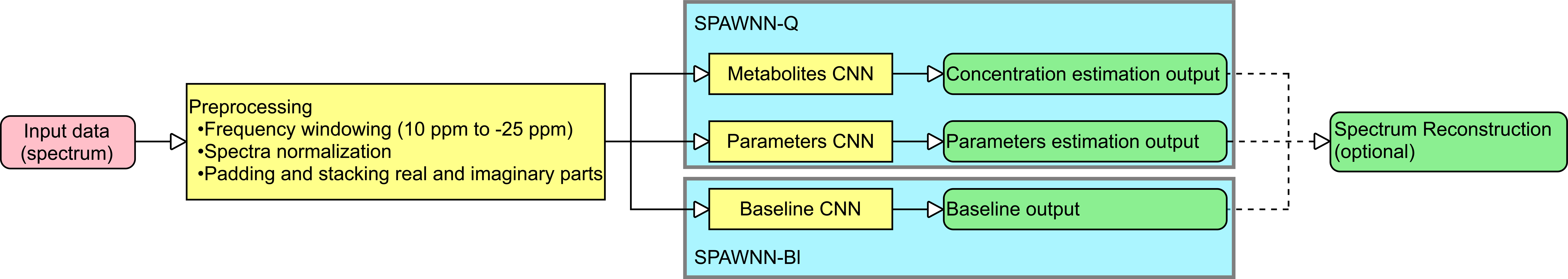
2. What questions did you ask yourselves while you were developing the code that would eventually be shared?
In the development phase, we aimed to create code that could be easily adapted and reused for different experiments or in different research scenarios.
We also considered the computational complexity and performance of our code to ensure that it could handle large training datasets or complex computations effectively.
Finally, we paid attention to the clarity and comprehensibility of our code. We aimed to write code that would be accessible to other researchers.
3. How do you recommend that people use the project repository you shared?
Start with the README file. Read the README file and try to rerun the command shown. it will provide an overview of the capability and usage of the code and model.
Once you have a good understanding of the code, try to run it on your own data, adapting the number of points and the bandwidth and retraining a new model.
Then, further adaptation could be implemented: you could add other metabolites, apply the method to other nuclei, explore other model architectures.
4. Are there any other reproducible research habits that you didn’t use for this paper but might be interested in trying in the future?
Using tools like Docker or Singularity to create reproducible and portable computing environments. This ensures that the code can be executed consistently across different systems and configurations without cumbersome installation or setup. In addition, keeping track of and archiving the successive versions of the code in a systematic manner allows researchers to easily track and manage changes to the code over time. This can be particularly useful when code is linked to a publication and extra data are required during the peer-review process. Also, we could try making automated pipelines that capture the entire workflow, from data acquisition to final results. Doing this simplifies the reproducibility process and reduces the potential for human error.




