Introduction
Quantitative MRI has a reproducibility problem. Even though we’ve been able to map fundamental MRI parameters (e.g. T1 relaxation) since the early days of MRI (Pykett and Mansfield 1978), forty years later measurements done using different quantitative techniques or by different groups can vary substantially (Stikov et al. 2015). Some techniques are commonly accepted as a gold standard (e.g. inversion recovery for T1), however differences in how these are implemented on scanners or how the data is processed may still impact the reproducibility of measured values across sites/studies. In the most recent edition of his book, Paul Tofts Paul Tofts introduced a medal system to describe a “perfect” quantitative MRI machine (i.e. technique) (Table 1).
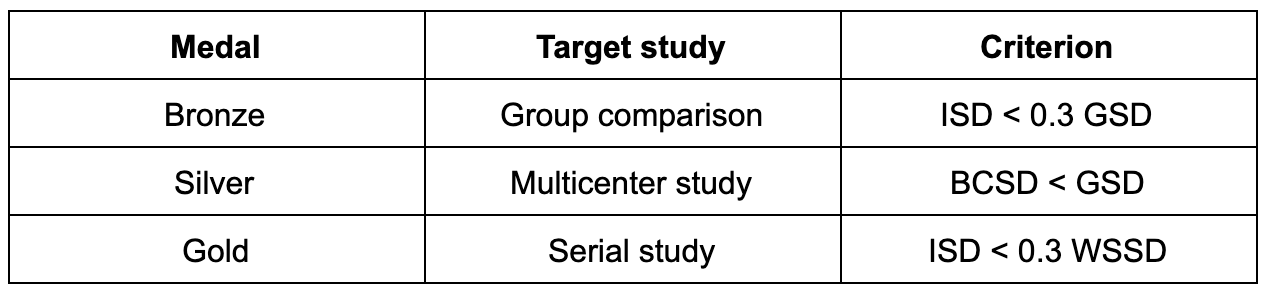
Table 1. Paul Tofts medal system for “perfect” quantitative MRI machines, reproduced from his book Quantitative MRI of the Brain (Wiley, 2nd Edition, Chapter 1, Table 3). Abbreviations: standard deviation (SD), instrumental SD (ISD), within-group SD (GSD), between-center SD (BCSD), within-subject SD (WSSD).
For this year’s challenge, we want to explore the topic of multicenter reproducibility of one of the fundamental MR parameters (T1), measured using the gold standard inversion recovery technique. To do this, we will use the robust methodology proposed by Barral et al. 2010, which presents a general fitting model that is insensitive to most pulse sequence effects. This methodology is based on the steady-state equations for the inversion recovery experiment, which doesn’t assume full signal recovery at the end of each repetition time or perfect flip angles. The authors also made their T1 fitting code available so that it may be reused. To avoid biological variations in this multi-site comparison, we propose that measurements be made on the ISMRM/NIST system phantom which has an array of fiducial spheres with different T1 values. We realize that some centers may not have access to this phantom, so we also encourage all participants to submit data from a healthy adult cohort.

To summarize, our challenge for this year poses the following question: will an imaging protocol independently-implemented at multiple centers reliably measure what is considered one of the fundamental MR parameters (T1) using the most robust technique (inversion recovery) in a standardized phantom (ISMRM/NIST system phantom). In addition, if enough submissions also measure some healthy human data, it may be possible to explore if this technique could possibly earn a silver medal according to Tofts medal system for a “perfect MRI machine”.
Plan
Material
For this challenge, the main focus will be on the ISMRM/NIST MRI System Phantom, which is currently available at many sites. It is a spherical liquid phantom containing arrays of spheres for different T1, T2, and proton density values, in addition to resolution insets and slice profile ramps. For the purpose of this challenge, data should be acquired using a single slice acquired in-plane with the T1-array (green spheres). Temperature must be recorded prior and after acquisition using the thermometer inside the phantom, and fitted T1 values will be calibrated using the recorded temperatures. Comprehensive instructions for how to set-up the phantom prior to scanning are described here.
In addition, we would like every participant to also scan at least one healthy adult subject with the same imaging protocol if possible. If you don’t have access to the ISMRM/NIST phantom, you may still participate in this challenge using only human data. Single-slice data should be acquired parallel to the AC-PC line, crossing the superior portion of the corpus callosum. Please follow your institutional ethics guidelines and consent procedures when scanning human participants. In addition, all data from challenges participants will be submitted to an open-data hosting website (www.osf.io), so a modification to your consent forms may be needed to inform your participants of this fact. Data must be anonymized prior to submission. This website has guidelines and examples on how to inform participants of open data sharing.
For consistency, and because T1 relaxation values depend on field strength, we recommend that data be acquired on a 3T clinical MRI system. However, data from any system/field strength is welcome as long as the pertinent hardware details are reported alongside the data.
Protocol
The recommended inversion recovery imaging protocol for this challenge is the one presented in Barral et al. 2010. It uses a stock spin-echo pulse sequence with a non-selective inversion preparation pulse. The main features are the following:
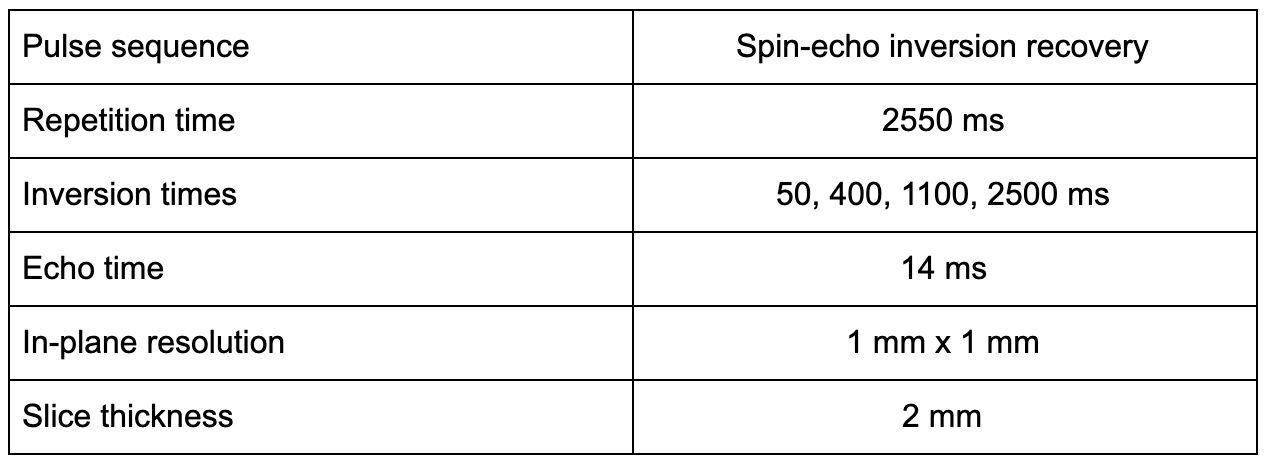
We ask that participants adhere to this protocol as best as possible, but if needed report any protocol differences they needed to make to accommodate the imaging sequence on their system. If possible, it is preferable that the data be exported in a complex data format (magnitude and phase, or real and imaginary), however the processing pipeline is compatible with magnitude-only data if the former is unavailable on your system.
Data Processing
All challenge data must be made openly accessible by uploading them to the OSF repository. To get your data uploaded, please contact one of our OSF data account administrators by opening an issue in the data registration GitHub repository. Together, you’ll follow a checklist to ensure that all required files are included, anonymized (if human), properly named, and correctly formatted. We ask that data be uploaded in the NIfTI format, with an accompanying JSON file containing all relevant information (a standard template will be provided, and can be expanded at your discretion).
A standardized T1 fitting script using Barral et al.’s original fitting code (using the qMRLab API for this package) will be made available in a repository of the GitHub organization for the challenge. This script will be made available as a Jupyter Notebook and accessible on MyBinder in a reproducible computing environment (thanks to Docker). Although participants may also submit T1 maps generated using their own fitting code, all image data will also be fit using the script mentioned above to standardize results from the challenge.
Once the T1 maps are all processed, a final analysis will be collaboratively developed on GitHub. We may reach out to a statistician to assist during this step, particularly if enough human data is submitted to determine if this technique qualifies for a silver qMRI medal.
Final Advice
The proposal we’ve developed here is only the minimum we are asking for the challenge. If you feel particularly motivated, we challenge you to go further and explore other avenues to study in addition to what we’ve laid out here (e.g. test-retest, a larger human sample cohort, different phantom temperatures, different inversion times, etc).
Lastly, note that the plan outlined here is meant to be a general guideline for you to follow. There is no need to reach out to check if you are doing a certain step “right”, unless you need some major clarifications about the challenge or to get more information about how to submit after the data is acquired. If to the best of your knowledge you believe you need to make an adjustment to the protocol for it to work for you, do it and report what you did differently. The main goal of this challenge is to explore how variable this particular method is to independent implementations across multiple centers, thus some variations in the protocol is expected.
Timeline
Registration deadline: January 17th 2020
Data must be uploaded by: February 29th 2020
Collaborative analysis will happen during the month of March 2020
Presentations during the ISMRM meeting in April 2020
How to Register
Please fill out this Google Form to register: https://forms.gle/bxLnHaWRyFJEH6Bp9
Frequently Asked Questions (FAQ)
I’m a bit rusty regarding the inversion recovery T1 mapping technique theory. Do you have any recommended resources/papers so that I can brush up on my theory?
This recently published interactive tutorial should cover most inversion recovery T1 mapping theory needed to understand this challenge: https://qmrlab.org/jekyll/2018/10/23/T1-mapping-inversion-recovery.html
It contains several interactive figures, and a MyBinder.org link to modify or rerun the code used to generate the blog post. Further resources are listed as references.
I don’t have access to the NIST phantom. Can I still participate?
One way you can still participate is to acquire human data with the recommended protocol and upload it anonymized. Another way you can participate if you can’t acquire data is to contribute to the GitHub repositories, where the code for the data processing and analysis will be worked on collaboratively.
I’m used to the simple three parameter inversion recovery equation (S = C(1+2exp(-TI/T1)), can I just fit my data with that instead?
That equations makes several assumptions (e.g. long TR, long delay between each TI and TR, ideal flip angles), and may not accurately fit the T1 values. For Barral’s method, the starting point is the full steady-state equation for the inversion recovery experiment and doesn’t make any further assumptions (i.e. it should fit accurately even for short TRs). If you want, you may compare the results by fitting your data with both equations/algorithms.
I noticed that some of your TIs are almost as long as your TR, leaving almost no time for signal recovery prior to the next inversion pulse. Isn’t that a problem in an inversion recovery experiment?
This misconception originates from the fact that the simple three parameter inversion recovery equation S = C(1+2exp(-TI/T1) assumes an experiment where the magnetization is fully recovered at the end of each TR. This case is what is commonly taught and referred to as the inversion recovery T1 mapping technique. However, a more robust approach is to consider the steady-state equation for this experiment (see Barral et al. 2010 and Gupta et al. 1980), which is compatible with TIs near the TR. This is what is done in the code made available by Barral et al., and is why this protocol can be used to measure T1 accurately. For further convincing, please refer the following interactive tutorial: https://qmrlab.org/jekyll/2018/10/23/T1-mapping-inversion-recovery.html
Who can I contact if I have questions about the challenge?
Please e-mail: rrsg.challenge@gmail.com
Acknowledgements
We would like to thank Paul Tofts, Joëlle Barral, Agâh Karakuzu, Julien Cohen-Adad and Ilana Leppert for valuable discussions and feedback on early versions of the challenge.
Reproducible Research Study Group Governing Committee
Martin Uecker (martin.uecker@med.uni-goettingen.de), Florian Knoll (florian.knoll@nyulangone.org), Nikola Stikov (nikola.stikov@polymtl.ca), Maria Eugenia Caligiuri (me.caligiuri@unicz.it), Daniel Gallichan (gallichand@cardiff.ac.uk)
Quantitative MR Study Group Governing Committee
Kathryn Keenan (kathryn.keenan@nist.gov), Diego Hernando (DHernando@uwhealth.org), Xavier Golay (x.golay@ucl.ac.uk), Annie Yuxin Zhang (yzhang785@wisc.edu), Jeff Gunter (Gunter.Jeffrey@mayo.edu)
Challenge leader: Mathieu Boudreau (Mathieu.Boudreau@icm-mhi.org)





