By Mathieu Boudreau
The February 2020 MRM Highlights interview is with Sebastiano Barbieri, Oliver Gurney-Champion, and Harriet Thoeny, researchers at UNSW Sydney in Australia, Amsterdam UMC in The Netherlands, and Hôpital Cantonal Fribourgois, University of Fribourg in Switzerland, respectively, and authors of a paper entitled “Deep learning how to fit an intravoxel incoherent motion model to diffusion‐weighted MRI”. Their paper was chosen as the MRM Highlights pick of the month because it reports reproducible research practices. In particular, in addition to sharing their code, the authors shared a demo of their code in a Jupyter Notebook document. We sent them a questionnaire in order to gain further insights into their reproducible research practices. However, before putting our questions to them, we wrote a brief “crash course” in order to get an overview of what Jupyter Notebook is all about. For more information about the authors and their research, check out our recent interview with them.
Crash Course: Jupyter Notebook

What is Jupyter Notebook? It is a browser-based coding environment that allows you to create shareable scripts as interactive documents. Code written in a Jupyter Notebook “notebook document” (or, more simply, a “Jupyter notebook”) is often accompanied by figures and text written in Markdown (simplified HTML) to guide the users through each executable cell. After execution, the notebook document can be exported in a variety of formats (e.g. iPython Notebook, HTML, PDF, slides, LaTeX, etc.). Jupyter notebooks can be helpful for a wide-range of applications in research; for providing a user-friendly demo of your software (as in this instance), for workshops and tutorials, for creating a self-contained record (commands, results, figures) of an analysis script, and so on. Jupyter notebooks are compatible with several other Project Jupyter initiatives, such as JupyterLab, Binder, and Jupyter Book. Other services are also compatible with Jupyter notebooks, such as Google Colab, Microsoft Azure Notebooks, and Code Ocean, although not all open-source. Jupyter Notebook plugins are compatible with these external services. You can test out the Jupyter Notebook environment in your web browser by clicking this link – no installation required.
General questions
1. Why did you choose to share your code/data?
We chose to share our code for several reasons:
- We wanted readers interested in the published paper to be able to gain a better understanding of the proposed algorithm by looking at the code.
- Sharing this information facilitates the reproducibility of the results.
- It allows researchers and interested clinicians to further improve the algorithm and test it on their own data, without spending valuable time re-implementing existing software.
2. Do you think you’ll share code/data again in future publications?
Certainly, for us, code and data sharing (e.g. through code repositories such as GitHub or data publications) is a priority wherever possible.
3. At what stage did you decide to share your code/data? Is there anything you wish you had known or done sooner?
We decided to share our code as soon as it became apparent that it was producing useful results. We are also in the process of publishing the medical images that were analyzed in our research, but this poses some additional legal and privacy-related challenges. We think that sharing of clinical data, in a way that respects patient privacy, should be encouraged, as it may limit the unnecessary duplication of clinical studies that collect similar data.
4. Are there any other reproducible research practices that you didn’t use for this paper but might be interested in trying in the future?
There is always room for improvement when documenting code and intermediate results. We also plan to facilitate access to the raw data underlying summary figures and tables.
Questions about your specific reproducible research practice
1. What practical advice do you have for people who would like to share parts of their research using Jupyter notebooks?
Perhaps it’s worth noting that Jupyter notebooks are not limited to Python code but support many other languages, such as R and Julia.
Editor’s note: With the proper plugin, you can also use multiple languages within a single notebook. For example, within the same Jupyter notebook you could process your data in MATLAB and then use a Python package such as Plotly to visualize the data.
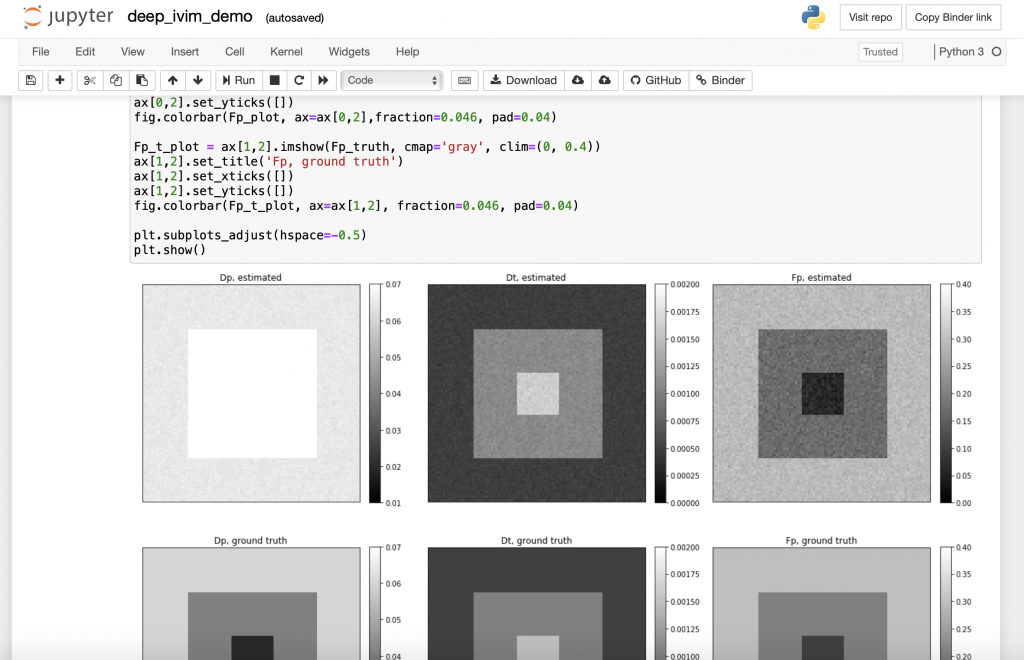
2. Can you share some resources that might help interested readers to get started using Jupyter Notebooks?
The Jupyter documentation is excellent (https://jupyter.org/documentation). For most first-time users it should be enough to take a look at the Markdown syntax (https://www.markdownguide.org/basic-syntax/) before formatting text in a notebook.
3. Did you encounter any challenges or hurdles during the process of writing or sharing a demo of your research using Jupyter notebooks?
To our knowledge, Jupyter notebooks shared on a GitHub repository cannot be run interactively without downloading them first*. Alternative options such as CloudStor SWAN or Google Colab offer this service and may be worth exploring in the future.
*Editor’s note: A recent open-source initiative by Project Jupyter called MyBinder now allows Jupyter notebooks hosted on GitHub to be run without the need to download them. All that is needed are some configuration files or a Dockerfile inside the GitHub repository. We forked the authors GitHub repository and added the necessary configuration file, you can now run their demo notebook in your web browser by simply clicking this link.
4. Besides being a useful tool to share your work with others, were there any other benefits of using Jupyter notebooks?
We found that the process of creating a Jupyter notebook helped us to identify and summarize the key elements of our research that may be of interest to fellow researchers and clinicians.




