By Mathieu Boudreau
The March 2020 MRM Highlights Reproducible Research Insights interview is with Shaihan Malik, a researcher from King’s College London, and lead author of a paper entitled “Steady‐state imaging with inhomogeneous magnetization transfer contrast using multiband radiofrequency pulses”. This paper was chosen as the MRM Highlights pick of the month because it reports good reproducible research practices. In particular, in addition to sharing their code, the authors produced exemplary code repository documentation (see: README.md), written using Markdown, a widely used text-to-HTML markup language. Shaihan’s research group takes a strong open-source stance, sharing code and data relating to their publications whenever possible, as shown by the repositories they have released on their @mriphysics GitHub organization. For more information about Shaihan Malik and his research, check out our recent interview with him and Daniel West, where we also got a bit carried away discussing open MRI science topics!
General questions
1. Why did you choose to share your code/data?
For a couple of reasons, the main one being that I’m a big believer in reproducible research. Much of the work in our field is in the form of complex calculations and requires software. It is not possible to accurately describe such methods in the brief conceptualised form of a paper, and hence there is a huge barrier to entry for any researcher wanting to reproduce or build on the results of a new technique. I think we should all be providing source code as an essential part of our publications since this allows methods to be reproduced. In addition, more selfishly, it is in the interests of all of us, as researchers, to enable others to use our methods – after all, more people using my methods increases the impact of my work!
2. Do you expect you’ll share code/data again in future publications?
Absolutely, everything that comes from our group is openly shared as a policy. This is also something that is currently pushed quite hard by the UK science funding agencies.
3. At what stage of this work did you decide to share your code/data? Is there anything you wish you had known or done sooner?
I’ve been doing this since setting up our GitHub site (@mriphysics) and we’ve shared a few things that have been used by others, including our EPG-X code for EPG simulation of exchanging systems – that work was a precursor to this research. The intention was to share this code right from the start of its development.
4. Are there any other reproducible research habits that you haven’t used for this paper that you might be interested in trying in the future?
We have pretty much stuck to sharing only code and non-human data. Sharing in vivo data is a bit more complex from the ethics perspective. For the type of work that we do, openly sharing source code is the main ‘reproducible research habit’ that I can think of. What others should we try?
Editor’s note: That’s a question that we hope our readers will continue to ask themselves. The purpose of this new MRM Highlights series on reproducible research practices is to highlight and contrast what “reproducible research practices” means for different groups. To see what other groups are doing, continue reading this series, and revisit our previous posts (the January post on sharing scripts for creating reproducible figures, and the February post on Jupyter notebooks).
Questions about the specific reproducible research habit

1. What advice do you have for people who would like to create good documentation on GitHub (i.e. README, Wiki, etc)?
I’m not sure I’m qualified to advise anyone here – certainly my approach to documentation isn’t anything like as sophisticated as a wiki (see here for an explanation of the differences between README files and wikis), which I would do if I could spare the time. As for the GitHub README, I think the key thing to have in mind is to make it readable. Try to explain everything from the perspective of someone who may not have read the paper (refer them to it) and be clear about how to *use* the code as well as understand it.
Editor’s note: The README file is also a great place to indicate how you’d like users to cite your work, as it’s typically the first and most visited page in your code repository.
2. Can you share some resources to help our readers get started with using Markdown?
To be honest I just googled every command before using it. 🙂
Editor’s note: So say we all! But to save readers a bit of time, here are two useful resources for getting started with Markdown.
- Syntax Guide for GitHub Markdown
- In-browser Markdown editor where you can view in real time what your Markdown commands look like.
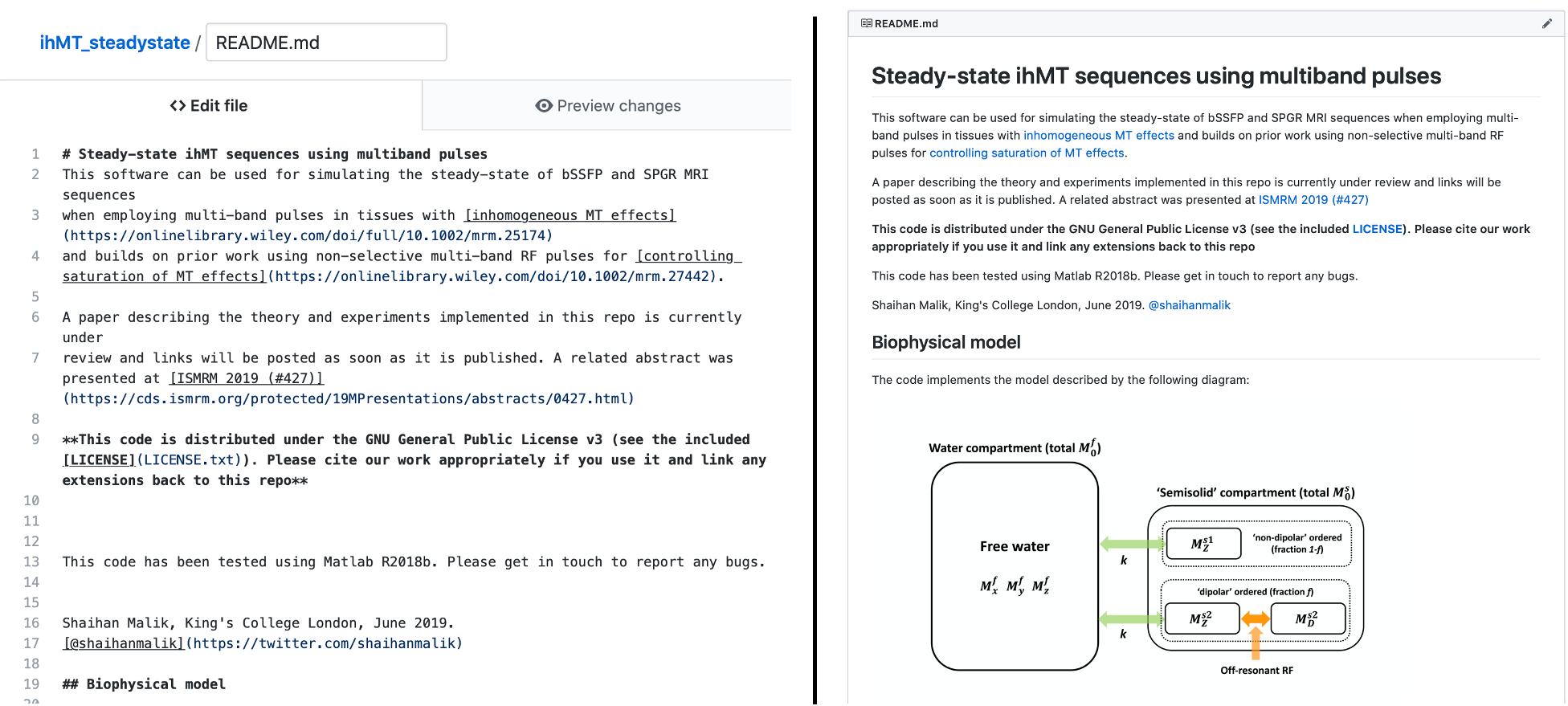
3. Have you noticed any other beneficial effects of having a well-documented README on Github?
The EPG-X code (which has been out longer) seems to have been used by quite a few people, some of whom have been in touch with me about it. In the end, your README is the best way to make your work accessible and hence improve its impact.
4. Is there anything else relating to reproducible research habits that you’d like to share with our readers?
It can be daunting to share code. Many people worry that things should be perfect before they are shared. But in my view, for many numerical (or image recon) methods, the code is an essential part of the ‘methods description’ of anything you publish. If you want to publish the results of something you should also be prepared to publish the method. So an approach I have found useful is to develop code with eventual open source publication in mind from the outset. Also, I guess I’m not so hung up on producing perfect code – far from it. 🙂




