By Mathieu Boudreau
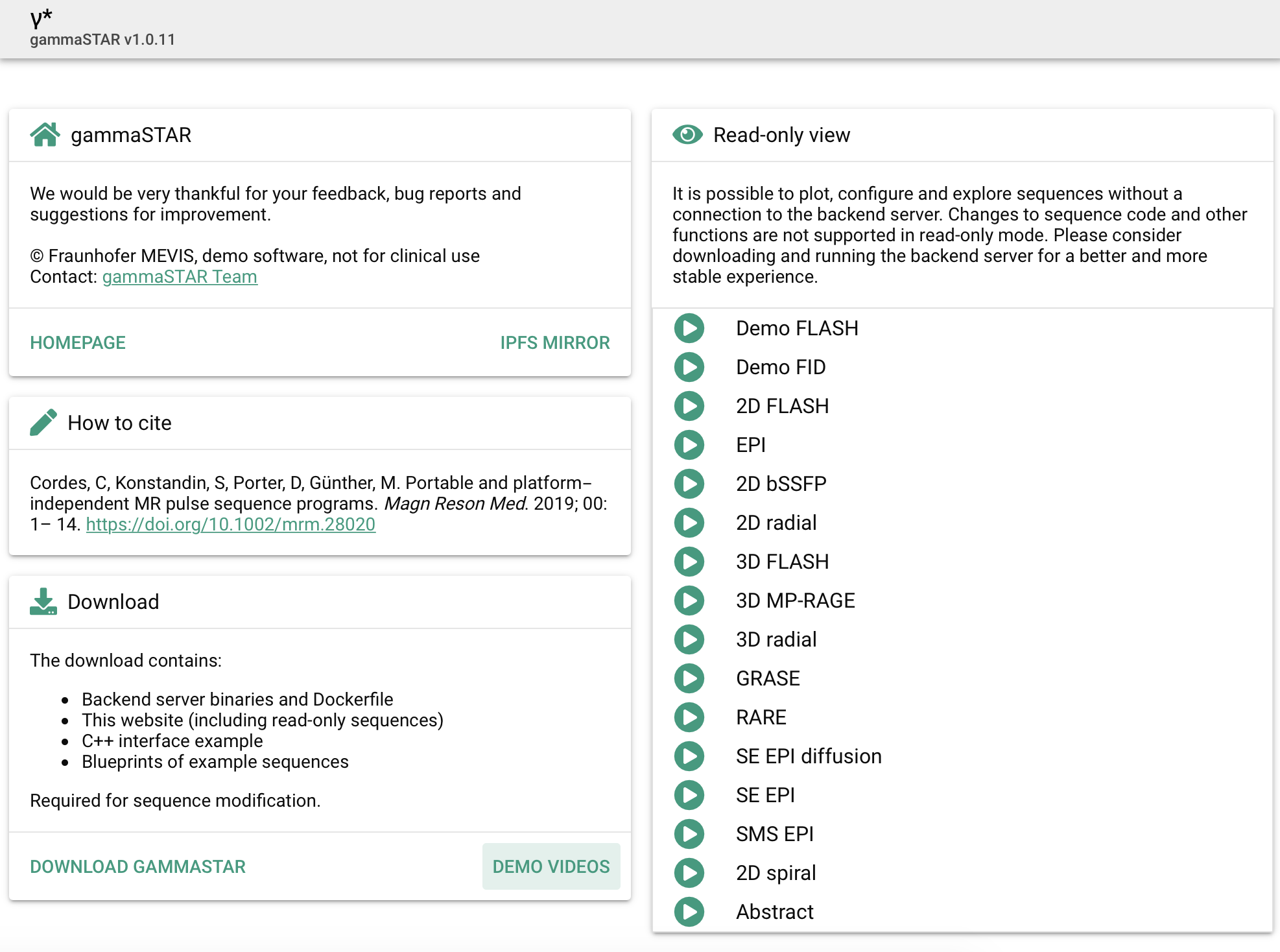
The May 2020 MRM Highlights Reproducible Research Insights interview is with Cristoffer Cordes and Matthias Günther, researchers at Fraunhofer Institute for Digital Medicine MEVIS in Bremen, Germany, and authors of a paper entitled “Portable and platform‐independent MR pulse sequence programs”. This paper was chosen as the MRM Highlights pick of the month because it reports good reproducible research practices. In particular, in addition to sharing their code for gammaSTAR, the authors also shared Dockerfiles enabling others to reproduce the coding environment needed to run their software. For more information about Cristoffer and Matthias and their research, check out our recent interview with them.
Crash Course: Docker
 Docker is a platform that allows you to create “containers”, a compilation of all the software, libraries, and tools needed to reproduce an environment to run your software. It’s like running a virtual machine for your software inside your computer or a server. The containers provide an environment that’s easily shareable and reproducible, and this makes them a great option to add to your reproducible research toolbox. You may have already encountered Docker containers without knowing it; services like MyBinder (covered in a previous MRM Highlights Reproducible Research blog post) use Docker containers to create and run environments for Jupyter Notebooks over the web. There are plenty of resources available to help you get started with Docker, and a collection of tools to simplify some of the steps (e.g. repo2docker).
Docker is a platform that allows you to create “containers”, a compilation of all the software, libraries, and tools needed to reproduce an environment to run your software. It’s like running a virtual machine for your software inside your computer or a server. The containers provide an environment that’s easily shareable and reproducible, and this makes them a great option to add to your reproducible research toolbox. You may have already encountered Docker containers without knowing it; services like MyBinder (covered in a previous MRM Highlights Reproducible Research blog post) use Docker containers to create and run environments for Jupyter Notebooks over the web. There are plenty of resources available to help you get started with Docker, and a collection of tools to simplify some of the steps (e.g. repo2docker).
General questions
1. Why did you choose to share your code/data?
Our software is a framework project. Frameworks grow best if they are used and experimented on, which is why we would like as many readers as possible to try out our software. We would also like users to scrutinize our work and gain a deeper understanding of our framework, because their feedback can help us fix some of its flaws and build on its advantages. Without sharing our code/data, readers would simply have to take our word for it when we describe subjective aspects of our framework, such as its flexibility, usability and performance. Without the possibility of reproducing its results, our claims would lose a lot of credibility.
2. Do you plan to share code/data regularly in future publications?
Yes, whenever possible.
3. At what stage of this work did you decide to share your code/data? Is there anything you wish you had known or done sooner?
It was clear from the beginning that there had to be some public version of the code/data. This can be tricky in a project that involves protected data that must not be shared publicly. Since we did not immediately decide on an exact strategy for separating public from protected code, doing this in the later stages was a tedious process.
4. Are there any other reproducible research habits that you didn’t use for this paper but might be interested in trying in the future?
The decentralized web could become more important in the future. Technologies such as the Interplanetary File System (IPFS) and blockchain might prove valuable assets in terms of reproducibility. Not relying on centralized service providers adds robustness and makes it easier for individuals to contribute. Furthermore, long-term archiving becomes simpler.
Questions about the specific reproducible research habits
1. What advice do you have for people who would like to develop a good user interface for their research software?
Most good contemporary user interfaces are realized using web technologies. It is not as hard as it may at first seem to move the UI to the web browser. That way, it is more accessible for users, easier to prototype, and the learning curve is very pleasant.
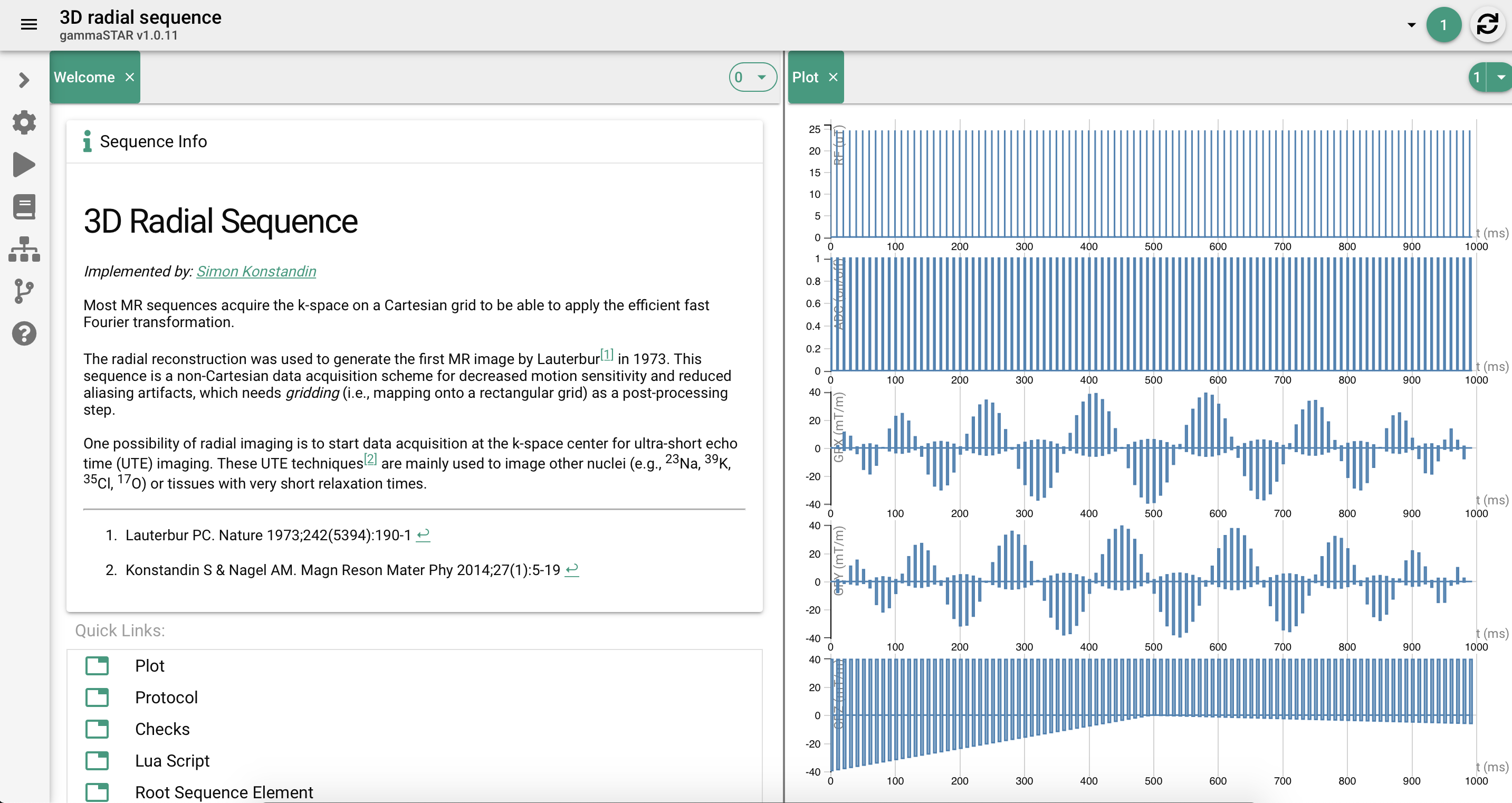
2. Why was it important for you to share a Dockerfile for this project? Can you share some resources to help our readers get started with using Docker containers?
Dockerfiles are among the easiest strategies to ensure that the correct software is installed the correct way and with as few commands as possible. It mostly removes the “but it runs fine on my machine” bias in a collaboration setting.
For anyone who wants to get started using Docker, we would recommend that before installing any software, they just do a Google search to see whether there is already a Docker image or Dockerfile available for their application. Docker images are hosted on Docker Hub, and many Dockerfiles are shared on GitHub. This is especially useful if your application involves databases or backend services.
For anyone who wants to use Docker for reproducible research, I would recommend that they start with a basic Docker image, write a Dockerfile to install the software that their project needs, then check if their project still works from within the container. After that, all that is needed for new users to get started with their project are convenience scripts.
3. Besides being a very useful resource for other researchers, what are the other benefits of having built a well-designed user interface for your software?
If users work with complex data, they need simplified views. If the simplified views leave open questions, then those need to be addressed as quickly and intuitively as possible. An interactive solution allows the user to choose what question is answered next.
A well-designed UI is useful not only for other researchers, but also for the developer, because a good overview of available information can make any problems with the software obvious and avoid the need for strenuous debugging efforts due to the developer being mostly blind to the state of the software.
4. Is there anything else relating to reproducible research habits that you’d like to share with our readers?
There are three key habits that we’d like to share:
- Never consider a part of a software “working” if it does not have a fully automatic test that can run on any developer’s machine. Features tend to break over time for various reasons, and problems tend to spread if they are not identified early.
- Setting up a developer’s machine should not require more than two manual steps. This feature helps other researchers reproduce your work without you having to spend a lot of time explaining all the steps, and it also helps you to keep in mind the prerequisites of your project
- Don’t forget to consider the implications of the software licenses you use. For example, GPL is very restrictive and its use by researchers is often prohibited; many researchers do not have a MATLAB license. Using such software will make your work unreproducible for many people.




