By Mathieu Boudreau
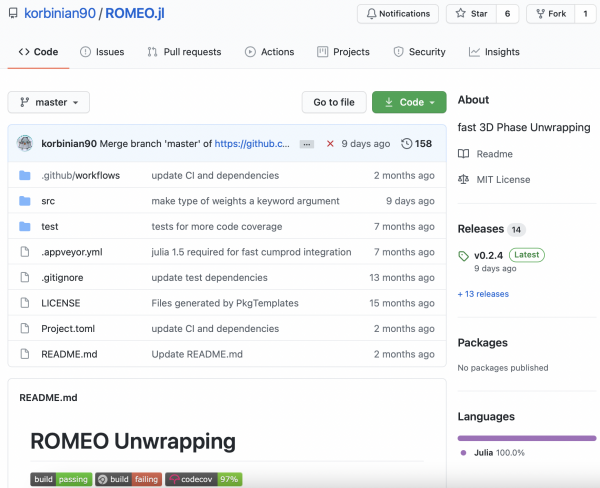
The March 2021 MRM Highlights Reproducible Research Insights interview is with Barbara Dymerska (University College London) and Korbinian Eckstein and Simon Robinson (Medical University of Vienna). Their paper, entitled Phase Unwrapping with a Rapid Opensource Minimum Spanning TreE AlgOrithm (ROMEO), introduces a new approach to phase unwrapping. It was chosen as this month’s Reproducible Research pick because the authors made the code for ROMEO open source and provided the community with executables for Windows, Linux and macOS. To learn more about these authors and their research, check out our recent interview with them.
To discuss this blog post, please visit our Discourse forum.
General questions
1. Why did you choose to share your code/data?
Simon: ROMEO is as much a tool as an algorithm: it would have been a travesty not to share it with the community after Korbinian put so much work into getting it to run efficiently. Also, I think that wherever it’s practicable, reviewers should be able to test a method and examine the results, rather than being obliged to try and assess images of selected slices in manuscript figures. Basically, attaching code bolsters a submission. Sharing also helps in dissemination: we were seeing enthusiastic tweets about ROMEO (with animated GIFs!) within hours of releasing it.
2. What is your lab or institutional policy on code/data sharing?
Simon: The Medical University of Vienna supports “FAIR Data Principles”; namely, that research data should be “findable, accessible, interoperable and reusable”. Making available the software and analysis scripts used to generate results is an essential part of that process. It is also the policy of The Austrian Science Fund, which funded this study, that data and software supporting publications should be accessible. For healthy subject data, we obviously anonymize the data, making sure all personal details are removed from image headers and titles. I see patient data as more problematic. We can ‘deface’ images to prevent facial recognition (Schwarz et al., 2019), but how do we guard against deanonymization methods that might be developed at some point in the future? And what about the rights to revoke consent and be forgotten (Politou et al., 2018)? We’re a long way off a dissemination system which makes that possible. That’s clear from current data hosting platforms, which are very basic. MRM’s preferred platform, XNAT, only hosts DICOM data, for instance, and in the Harvard Database, which we opted for in this project, it’s not even possible to create folders. These systems are a long way off being future-proof.
Barbara: Sharing data is great but we must do it responsibly. At University College London we have to do Data Protection Online training. As Simon mentioned, there are already facial recognition algorithms that can identify people from MRI scans and breach personal privacy. Most certainly, the future will bring even more powerful algorithms. Anonymous synthetic data generated using advanced machine learning algorithms is a solution that has recently caught my eye (Reiner Benaim et al., 2020). Just to avoid confusion, I am not talking about synthetic MRI as in (Riederer et al., 1986), where, from a single quantitative MRI sequence, images with different contrast are derived, all of which carry features specific for a given volunteer. Truly anonymous synthetic data would look realistic (like the photos of AI-generated people) and carry statistically relevant information, but would be completely depersonalized. This solution promises strong privacy protection along with the freedom to share big data for research and development purposes. There is currently no toolbox for MRI data able to create realistic anonymized synthetic MRI images. Dear MRI community – are you interested?
3. At what stage did you decide to share your code/data? Is there anything you wish you had known or done sooner?
Simon: We worked on this project remotely from an early (pre-pandemic) stage, so we thought about how to share data and code from the very outset. Barbara had already moved from Vienna to UCL by the time she became involved, and Korbinian and I both moved while we were working on it, Korbinian to Tübingen and I to Brisbane, so we’ve been using the data and code repositories ourselves as we’ve gone through the testing, writing and revising stages. I’m sure an increase in working from home during 2021 is having the same effect in other groups; collaborating on code development using github and gitlab and running it on data held on institutional cloud storage. This approach encourages us to keep things tidy and well commented, and therefore makes it easier to share more widely on submission or publication.
Korbinian: Code sharing started when Barbara joined the ROMEO team. The first steps consisted of copying scripts, which was tedious and involved setting up the same environment with all dependencies. However, the Julia package manager supports dependency files and can handle the whole setup process. Then it was only a small step to turn ROMEO into a package that can be easily shared. I think it would be beneficial to start a new codebase immediately with the idea of working together in a team. This requires that the project is designed for reproducibility from the start, an approach that leads to better code quality overall. The goal in a cooperative project is to write the code in such a way that someone else can read, understand and execute it.
If I were starting over with this project, I would begin writing tests as early as possible. It doesn’t take much effort to write simple tests for a newly implemented feature, and it is a lot easier to develop software with existing tests, as you get alerted when you accidentally break some existing behavior. Adding tests later on is much more tedious and does little to facilitate the development.
4. How do you think we might encourage researchers in the MRI community to contribute more open-source code along with their research papers?
Simon: These Highlights features are definitely a source of encouragement, as is the information included in the “Data Availability” section of the MRM Author Guidelines. Handling editors and reviewers can also suggest that authors share code, if that would help the review or increase a paper’s impact. Funding agencies are starting to demand this, too, so I’m optimistic that we will get there soon.
Questions about the specific reproducible research habit
1. Why did you choose to write your tool in Julia?
Korbinian: ROMEO started life as a simple project. My aim was to try out the new Julia programming language after we saw an ISMRM abstract where the method was written in Julia. As the syntax is superficially similar, it was easy to transition from MATLAB to Julia. It turned out to be the perfect language for the project as it is compiled (it can be as fast as C), and it is possible to implement the low-level data structures that we required for ROMEO (bucket queue to have a runtime of O(n)). Now, after finishing the project, I’m even more convinced Julia was the right choice. The Julia ecosystem encourages the use of structured and versioned open-source repositories, which can be created automatically with a simple customizable command1, optionally setting up CI/CD templates for writing tests, templates for hosting documentation, and so on). There is also a straightforward way to create an executable that users can run without installing Julia2.
2. What advice do you have for people who would like to write tests for their code?
Korbinian: Often, scientific code is changed as little as possible in order to avoid the risk of breaking an existing behavior. Therefore, an inefficient or ugly design is often kept, while new features make the program even harder to understand. I think software tests can solve this problem, greatly improve the development efficiency, and also increase the enjoyment of the development process. Because with tests, anyone involved in the project can add features or modify the code without the fear of breaking things. To achieve this goal, tests should be low-effort to write so as not to hinder the development process, and also low-effort to run for any change that might have unwanted effects. Therefore, my suggestions for tests would be: they need to run fast and also cover most of the codebase; make sure you test the program from the user perspective to verify the operation of certain features that shouldn’t change; don’t directly test internal code, which might change often, but instead test outside behavior as this verifies the functionality and implicitly tests the internal code; for data processing code (like ROMEO), test some simple metrics on small real data sets; finally, a simple tip: after fixing a bug, write a test that ensures that this bug cannot occur again — this will also give you a good feeling of having made an important contribution. Creating a good test setup is very important and most languages provide good test packages for automated testing. Online repositories also have the possibility to automatically run the tests for each git commit and show a small green badge if all tests succeed; that way, potential users will know that the code is working properly.
Here, I have focused only on tests that ensure an efficient development process, but depending on the project, there might be additional testing requirements.
Barbara: Choose challenging but realistic input data, which should test your code at extremes but also be close enough to the data you might expect to find in the database of the target MRI community (e.g., the QSM or MR thermometry communities). In our case, this meant including simulated complex topographies (with and without noise), and gradient echo and echo planar imaging data from high and ultra-high field MRI scanners: 3, 7 and 9.4 Tesla. I included inputs, as well as expected results, an approach that allows potential users to compare their outcomes with yours.
Remember to test every parallel option/feature you offer, even if you are convinced the defaults are the best. After all, what is the point of offering an alternative if it does not work as it should?
In MRI we are dealing with images: visual inspection of every major step of your algorithm is helpful in reaching a better understanding of the code behavior, facilitating better test design as well as code improvement. In our case, visualization of various ROMEO weights helped me to understand their interplay and relative importance in anatomical features such as veins or brain boundaries, as well as in pure noise regions.
Finally, ask people in your group to test the code on both your data and their own, and come back to you with their feedback — it can be surprising, and rewarding.
3. What questions did you ask yourselves when preparing your code for sharing?
Korbinian: We asked ourselves how we could make the application of ROMEO as simple and straightforward as possible, and decided to release a command line tool together with the source code. The next question was how to make it run on all major operating systems. The Julia code itself runs on any operating system, but the command line version of our software had to be specifically compiled for each one. Also, the open-source software license is important, as otherwise no one is allowed to use the code. We decided to use the MIT license, which is the least restrictive in terms of possible usages of ROMEO.
4. Are there any other reproducible research habits that you haven’t used for this paper that you might be interested in trying in the future?
Simon: We’re aware that we’re far from the cutting edge of the reproducibility movement. Singularity and Docker containers allow entire environments to be reproduced and shared, regardless of the user’s host operating system (though not necessarily the hardware, if software is compiled with hardware-specific optimization). Containers make sense for multi-step analyses involving lots of different software packages, which are more typical of neuroimaging studies, and would probably be overkill for ROMEO, which doesn’t have complex dependencies, but they are something we could well use in a more applied study. Beyond that, we are keeping an eye on the evolving NeuroDesk project3,4. NeuroDesk provides an ecosystem for making reproducible and scalable neuroimaging software containers for all operating systems and high-performance computing systems.
Barbara: I’ve used Jupyter Notebooks in Google Colab, which is great for undergrad projects, mainly as it is straightforward to use and you do not need to perform any local setup. I would be interested to test similar cloud services such as Kaggle and Azure Notebooks, especially for working in a team.
References
Politou, E., Alepis, E., & Patsakis, C. (2018). Forgetting personal data and revoking consent under the GDPR: Challenges and proposed solutions. Journal of Cybersecurity, 4(1). https://doi.org/10.1093/cybsec/tyy001
Reiner Benaim, A., Almog, R., Gorelik, Y., Hochberg, I., Nassar, L., Mashiach, T., Khamaisi, M., Lurie, Y., Azzam, Z. S., Khoury, J., Kurnik, D., & Beyar, R. (2020). Analyzing Medical Research Results Based on Synthetic Data and Their Relation to Real Data Results: Systematic Comparison From Five Observational Studies. JMIR Medical Informatics, 8(2), e16492.
Riederer, S. J., Lee, J. N., Farzaneh, F., Wang, H. Z., & Wright, R. C. (1986). Magnetic resonance image synthesis. Clinical implementation. Acta Radiologica. Supplementum, 369, 466–468.
Schwarz, C. G., Kremers, W. K., Therneau, T. M., Sharp, R. R., Gunter, J. L., Vemuri, P., Arani, A., Spychalla, A. J., Kantarci, K., Knopman, D. S., Petersen, R. C., & Jack, C. R., Jr. (2019). Identification of Anonymous MRI Research Participants with Face-Recognition Software. The New England Journal of Medicine, 381(17), 1684–1686.




