By Mathieu Boudreau
This MRM Highlights Reproducible Research Insights interview is with Jakob Assländer from NYU Langone Health, author, with others, of a paper entitled “Generalized Bloch model: A theory for pulsed magnetization transfer”. We chose to highlight this paper because the authors demonstrated exemplary reproducible research practices; in particular, they shared all the code, data, and scripts needed to reproduce their figures, and also created a beautiful tutorial with interactive figures and MyBinder links to run the code from any browser. To learn more, check out our recent interview with Jakob Assländer and Daniel Sodickson.
To discuss this feature, please visit our Discourse forum.
General questions
1. Why did you choose to share your code/data?
Being funded with public money, I feel it is my job to facilitate scientific progress. In my humble opinion, sharing publishing code and data is an important part of working together as a scientific community. But I think there are also benefits to be had for authors, too, as open source code can help to promote the publications that relate to it.
2. What is your lab or institutional policy on code/data sharing?
Our department is putting a lot of effort into curating and sharing code, data, and even hardware. Our shared resources, available through the Center for Advanced Imaging Innovation and Research, include over 50 software packages that have been downloaded over 15,000 times. The fastMRI dataset contains the raw data of over 8000 scans and the download traffic of this dataset is about 1000 TB/year. Furthermore, we are shipping, free of charge, a pilot tone transmitter to interested academic sites. Long story short, I could not wish for an environment that places greater emphasis on sharing code and data.
3. At what stage did you decide to share your code/data? Is there anything you wish you had known or done sooner?

To me, deciding to write a paper about a topic and choosing to publish any useful code associated with it are one and the same thing. To answer the second question, with every software project, I feel that I start cleaning up the code and writing tests too late. Both really can make programming much more efficient, and so these steps should be taken early on.
4. How do you think we might encourage researchers in the MRI community to contribute more open-source code along with their research papers?
My experience has always been that publishing code helped me more than it hurt me. Hopefully, this blog, filled with many success stories, will help!
Questions about the specific reproducible research habit
1. Why did you choose to do your work with the Julia programming language?
That is a great question and I am very opinionated about this: I think Julia is the future of scientific computing and that we should all switch to it! It allows you to jot down an idea with the same ease as MATLAB and Python, and combines this with C-like speed, all in the same language! In addition, the ecosystem, the package manager, and available IDEs are all state-of-the-art and very thought through, making it easy to use them, easy to share code, and easy to reproduce the same environment.
2. What advice do you have for creating reproducible figures?
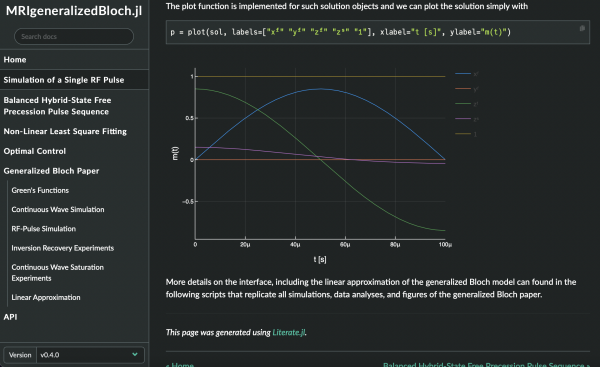
Not sure I have advice on this. I create my figures in LaTeX and since I upload the whole thing on ArXiv, they are, I guess, reproducible and hackable. But figures in LaTeX are a somewhat painful process, so I am not sure I can give a whole-hearted recommendation.
3. As a way of showcasing the research from your article, you’ve created a beautiful website that incorporates code, interactive figures, and text. What motivated you to put in the extra effort in making it, and do you have any tips on using the tools you needed to create it?
I must say that the Julia packages Documenter.jl and Literate.jl made this a lot easier. But still, I can’t deny that it takes some work to set things up and create the content. I think I was just hoping that people would find it useful, and maybe it will inspire others to replicate this approach. From a technological point of view, it is very easy to replicate: everything is open source and published in my GitHub repository, so it is just a matter of cloning the repository and replacing the content.
4. Are there any other reproducible research habits that you didn’t use for this paper but might be interested in trying in the future?
In this project I only used a very small NMR dataset, which I simply published in a GitHub repository. One day, I will have to learn how best to publish larger MRI datasets and how to deal with facial recognition from high-resolution 3D datasets.




